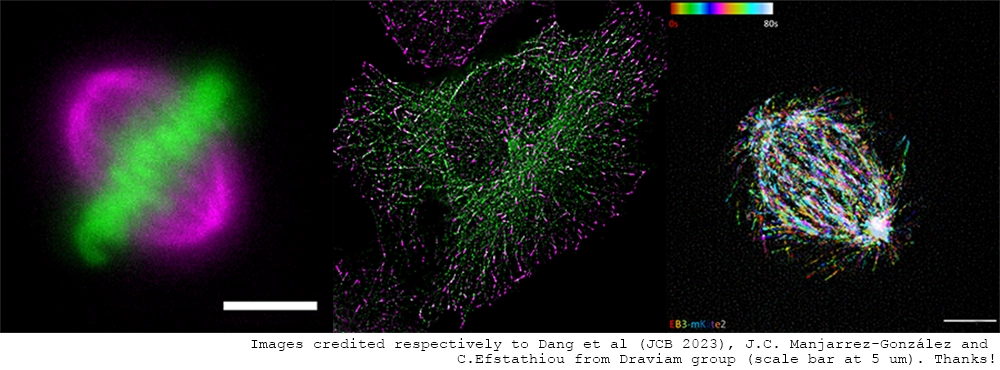
Cell Dynamics And Chromosomal Stability Workshop
We will hold a workshop for the topic of cell division and migration dynamics and the chromosome segregation control during cell proliferation, with presentations about SpinX, the supported studies by Carl Zeiss, which uses Deep learning techniques for 3D + time series images. Established scientists in this field will be gathered here and give special talks.
No registration fee to join, we requested you to register you name and address below.
Registration Form is Here (or Here if you find difficult to access Google Form)
Programme
13:00 Desk open for the workshop and the poster session. Coffee and Sweets will be served around the registration desk.
13:20 Welcome opening remarks and Keynote speaker short introduction (Prof. Jingyan Fu & Prof. Viji M. Draviam)
13:30 Prof. Viji M. Draviam - Chromosome capture mechanisms that ensure chromosomal stability (40 minutes + 5 minutes discussion)
14:20 Prof. Jingyan Fu - Protein architecture at drosophila centriole core (40 minutes + 5 minutes discussion)
15:10 Short break - Coffee/Tea/Sweets (10 minutes)
15:20 Dr. Binghao Chai - AI in biomedical image and movie analysis (30 minutes + 5 minutes discussion)
16:00 Jinwang Lu - Cell segmentation in low signal-to-noise ratio microscopy images based on machine learning (15 minutes + 5 minutes discussion)
16:20 Dr Denise Ragusa - Contribution of histone variants to chromosome instability: focus on H2A.Z variants (15 minutes + 5 minutes discussion)
16:45 Dr. Binghao Chai - Hands-on AI guided image analysis workshop. Showcase of SpinX and nuclear tracker on ZEISS arivis Cloud (formerly APEER) (60 minutes)
17:45-17:55 Closing remarks of workshop (Jingyan and Viji, Thank-you note).